MIGEC: Molecular Identifier Guided Error Correction pipeline¶
FEATURES¶
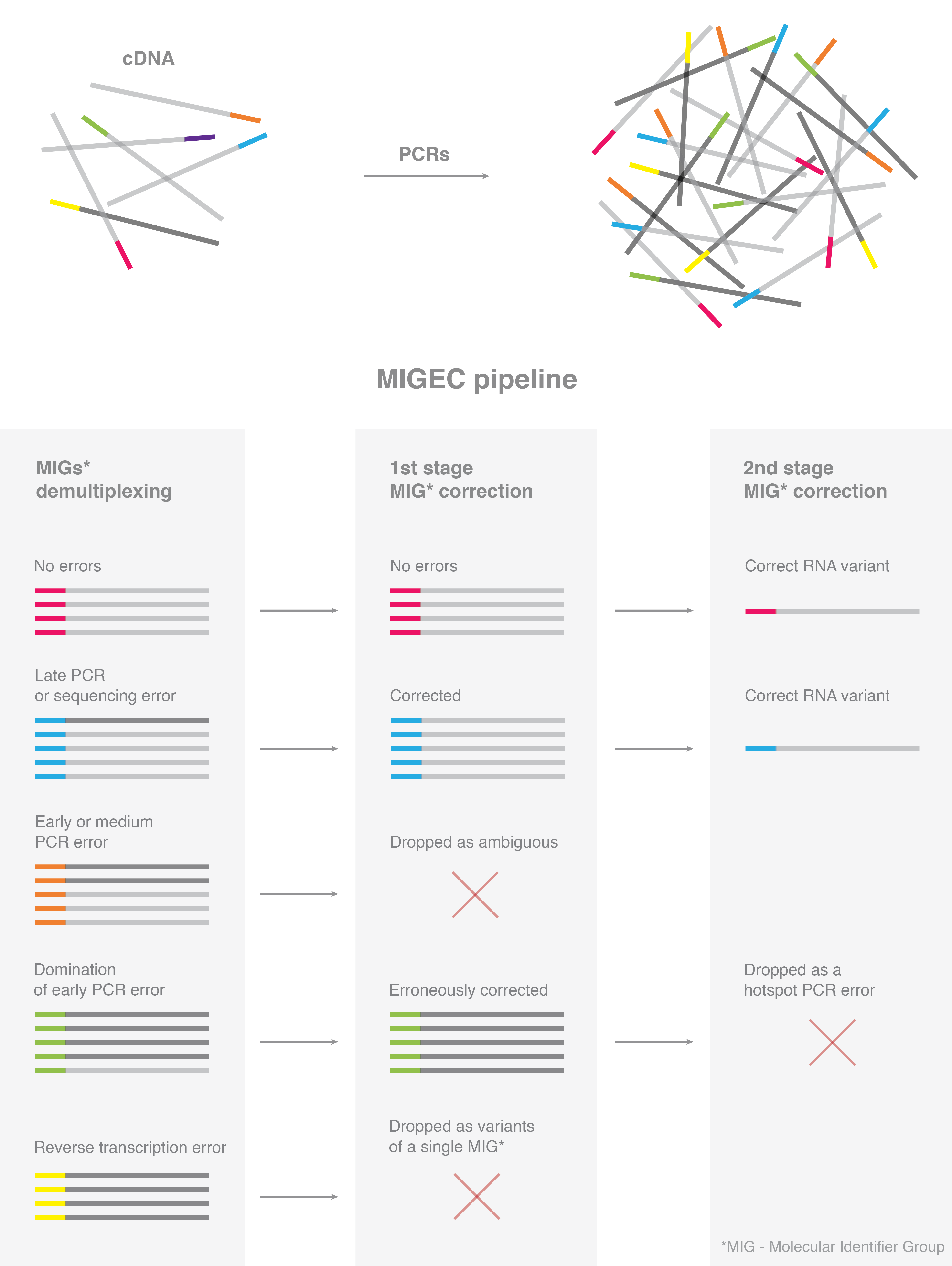
- De-multiplexing, adapter trimming and read overlapping for NGS data. Extraction of UMI sequences
- Assembly of consensuses for original molecules that entered library preparation by grouping reads with identical molecular identifiers
- Mapping of V, D and J segments, extraction of CDR3 regions and clonotype assembly for all human and mouse immune receptors (TRA/TRB/TRG/TRD and IGH/IGL/IGK)
- Additional filtering of hot-spot errors in CDR3 sequence
- Flexible and straightforward batch processing
- Currently all species-gene pairs that have germline sequences (based on IMGT) that allow CDR3 identification are supported
| Species | Gene |
|---|---|
| HomoSapiens | TRA, TRB, TRG, TRD, IGL, IGK, IGH |
| MusMusculus | TRB, TRG, TRD, IGL, IGK, IGH |
| MacacaMulatta | TRB, IGK, IGH |
| OryctolagusCuniculus | IGL, IGK, IGH |
| RattusNorvegicus | IGL, IGH |
| CanisLupusFamiliaris | TRB, TRG |
| SusScrofa | IGL, IGK |
| BosTaurus | TRD |
| MusSpretus | IGL |
| GallusGallus | TRB |
| AnasPlatyrhynchos | TRB |